您好,登錄后才能下訂單哦!
您好,登錄后才能下訂單哦!
這篇文章主要講解了用python進行時間序列分析的方法,內容清晰明了,對此有興趣的小伙伴可以學習一下,相信大家閱讀完之后會有幫助。
什么是時間序列
時間序列簡單的說就是各時間點上形成的數值序列,時間序列分析就是通過觀察歷史數據預測未來的值。在這里需要強調一點的是,時間序列分析并不是關于時間的回歸,它主要是研究自身的變化規律的(這里不考慮含外生變量的時間序列)。
為什么用python
用兩個字總結“情懷”,愛屋及烏,個人比較喜歡python,就用python擼了。能做時間序列的軟件很多,SAS、R、SPSS、Eviews甚至matlab等等,實際工作中應用得比較多的應該還是SAS和R,前者推薦王燕寫的《應用時間序列分析》,后者推薦“基于R語言的時間序列建模完整教程”這篇博文(翻譯版)。python作為科學計算的利器,當然也有相關分析的包:statsmodels中tsa模塊,當然這個包和SAS、R是比不了,但是python有另一個神器:pandas!pandas在時間序列上的應用,能簡化我們很多的工作。
環境配置
python推薦直接裝Anaconda,它集成了許多科學計算包,有一些包自己手動去裝還是挺費勁的。statsmodels需要自己去安裝,這里我推薦使用0.6的穩定版,0.7及其以上的版本能在github上找到,該版本在安裝時會用C編譯好,所以修改底層的一些代碼將不會起作用。
時間序列分析
1.基本模型
自回歸移動平均模型(ARMA(p,q))是時間序列中最為重要的模型之一,它主要由兩部分組成: AR代表p階自回歸過程,MA代表q階移動平均過程,其公式如下:
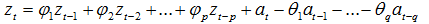
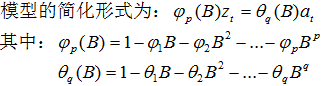
依據模型的形式、特性及自相關和偏自相關函數的特征,總結如下:
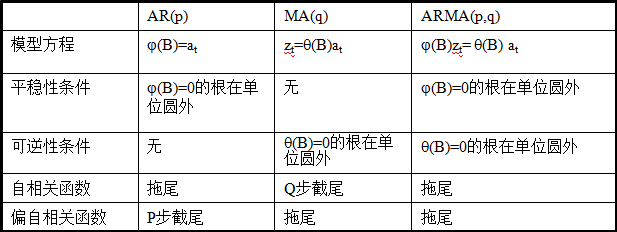
在時間序列中,ARIMA模型是在ARMA模型的基礎上多了差分的操作。
2.pandas時間序列操作
大熊貓真的很可愛,這里簡單介紹一下它在時間序列上的可愛之處。和許多時間序列分析一樣,本文同樣使用航空乘客數據(AirPassengers.csv)作為樣例。
數據讀取:
# -*- coding:utf-8 -*-
import numpy as np
import pandas as pdfrom datetime import datetimeimport matplotlib.pylab as plt
# 讀取數據,pd.read_csv默認生成DataFrame對象,需將其轉換成Series對象df = pd.read_csv('AirPassengers.csv', encoding='utf-8', index_col='date')df.index = pd.to_datetime(df.index) # 將字符串索引轉換成時間索引ts = df['x'] # 生成pd.Series對象# 查看數據格式ts.head()ts.head().index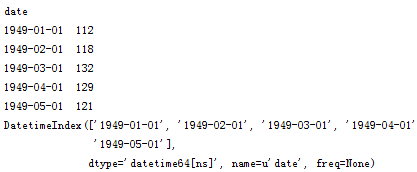
查看某日的值既可以使用字符串作為索引,又可以直接使用時間對象作為索引
兩者的返回值都是第一個序列值:112
如果要查看某一年的數據,pandas也能非常方便的實現
ts['1949']

切片操作:
ts['1949-1' : '1949-6']
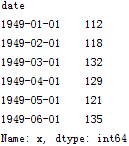
注意時間索引的切片操作起點和尾部都是包含的,這點與數值索引有所不同
pandas還有很多方便的時間序列函數,在后面的實際應用中在進行說明。
3. 平穩性檢驗
我們知道序列平穩性是進行時間序列分析的前提條件,很多人都會有疑問,為什么要滿足平穩性的要求呢?在大數定理和中心定理中要求樣本同分布(這里同分布等價于時間序列中的平穩性),而我們的建模過程中有很多都是建立在大數定理和中心極限定理的前提條件下的,如果它不滿足,得到的許多結論都是不可靠的。以虛假回歸為例,當響應變量和輸入變量都平穩時,我們用t統計量檢驗標準化系數的顯著性。而當響應變量和輸入變量不平穩時,其標準化系數不在滿足t分布,這時再用t檢驗來進行顯著性分析,導致拒絕原假設的概率增加,即容易犯第一類錯誤,從而得出錯誤的結論。
平穩時間序列有兩種定義:嚴平穩和寬平穩
嚴平穩顧名思義,是一種條件非常苛刻的平穩性,它要求序列隨著時間的推移,其統計性質保持不變。對于任意的τ,其聯合概率密度函數滿足:
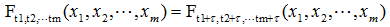
嚴平穩的條件只是理論上的存在,現實中用得比較多的是寬平穩的條件。
寬平穩也叫弱平穩或者二階平穩(均值和方差平穩),它應滿足:
平穩性檢驗:觀察法和單位根檢驗法
基于此,我寫了一個名為test_stationarity的統計性檢驗模塊,以便將某些統計檢驗結果更加直觀的展現出來。
# -*- coding:utf-8 -*-
from statsmodels.tsa.stattools import adfuller
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
# 移動平均圖
def draw_trend(timeSeries, size):
f = plt.figure(facecolor='white')
# 對size個數據進行移動平均
rol_mean = timeSeries.rolling(window=size).mean()
# 對size個數據進行加權移動平均
rol_weighted_mean = pd.ewma(timeSeries, span=size)
timeSeries.plot(color='blue', label='Original')
rolmean.plot(color='red', label='Rolling Mean')
rol_weighted_mean.plot(color='black', label='Weighted Rolling Mean')
plt.legend(loc='best')
plt.title('Rolling Mean')
plt.show()
def draw_ts(timeSeries): f = plt.figure(facecolor='white')
timeSeries.plot(color='blue')
plt.show()
''' Unit Root Test
The null hypothesis of the Augmented Dickey-Fuller is that there is a unit
root, with the alternative that there is no unit root. That is to say the
bigger the p-value the more reason we assert that there is a unit root
'''
def testStationarity(ts):
dftest = adfuller(ts)
# 對上述函數求得的值進行語義描述
dfoutput = pd.Series(dftest[0:4], index=['Test Statistic','p-value','#Lags Used','Number of Observations Used'])
for key,value in dftest[4].items():
dfoutput['Critical Value (%s)'%key] = value
return dfoutput
# 自相關和偏相關圖,默認階數為31階
def draw_acf_pacf(ts, lags=31):
f = plt.figure(facecolor='white')
ax1 = f.add_subplot(211)
plot_acf(ts, lags=31, ax=ax1)
ax2 = f.add_subplot(212)
plot_pacf(ts, lags=31, ax=ax2)
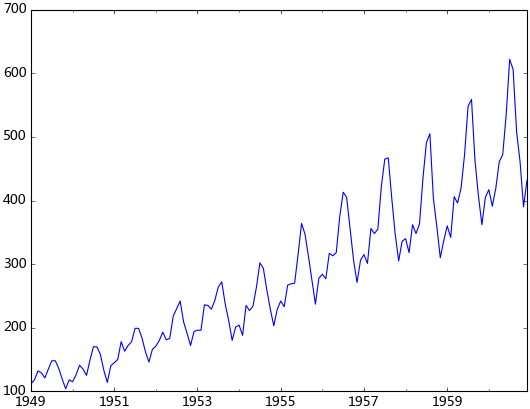
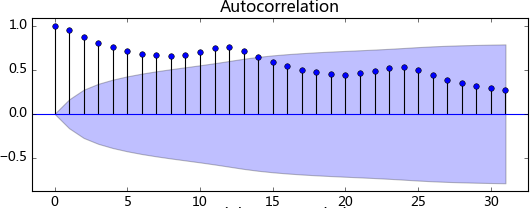
plt.show()觀察法,通俗的說就是通過觀察序列的趨勢圖與相關圖是否隨著時間的變化呈現出某種規律。所謂的規律就是時間序列經常提到的周期性因素,現實中遇到得比較多的是線性周期成分,這類周期成分可以采用差分或者移動平均來解決,而對于非線性周期成分的處理相對比較復雜,需要采用某些分解的方法。下圖為航空數據的線性圖,可以明顯的看出它具有年周期成分和長期趨勢成分。平穩序列的自相關系數會快速衰減,下面的自相關圖并不能體現出該特征,所以我們有理由相信該序列是不平穩的。
單位根檢驗:ADF是一種常用的單位根檢驗方法,他的原假設為序列具有單位根,即非平穩,對于一個平穩的時序數據,就需要在給定的置信水平上顯著,拒絕原假設。ADF只是單位根檢驗的方法之一,如果想采用其他檢驗方法,可以安裝第三方包arch,里面提供了更加全面的單位根檢驗方法,個人還是比較鐘情ADF檢驗。以下為檢驗結果,其p值大于0.99,說明并不能拒絕原假設。

3. 平穩性處理
由前面的分析可知,該序列是不平穩的,然而平穩性是時間序列分析的前提條件,故我們需要對不平穩的序列進行處理將其轉換成平穩的序列。
a. 對數變換
對數變換主要是為了減小數據的振動幅度,使其線性規律更加明顯(我是這么理解的時間序列模型大部分都是線性的,為了盡量降低非線性的因素,需要對其進行預處理,也許我理解的不對)。對數變換相當于增加了一個懲罰機制,數據越大其懲罰越大,數據越小懲罰越小。這里強調一下,變換的序列需要滿足大于0,小于0的數據不存在對數變換。
ts_log = np.log(ts) test_stationarity.draw_ts(ts_log)
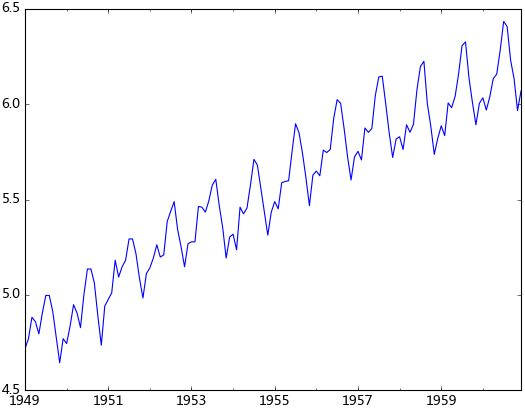
b. 平滑法
根據平滑技術的不同,平滑法具體分為移動平均法和指數平均法。
移動平均即利用一定時間間隔內的平均值作為某一期的估計值,而指數平均則是用變權的方法來計算均值
test_stationarity.draw_trend(ts_log, 12)
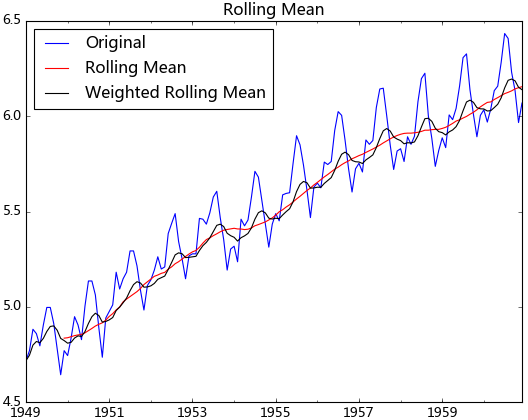
從上圖可以發現窗口為12的移動平均能較好的剔除年周期性因素,而指數平均法是對周期內的數據進行了加權,能在一定程度上減小年周期因素,但并不能完全剔除,如要完全剔除可以進一步進行差分操作。
c. 差分
時間序列最常用來剔除周期性因素的方法當屬差分了,它主要是對等周期間隔的數據進行線性求減。前面我們說過,ARIMA模型相對ARMA模型,僅多了差分操作,ARIMA模型幾乎是所有時間序列軟件都支持的,差分的實現與還原都非常方便。而statsmodel中,對差分的支持不是很好,它不支持高階和多階差分,為什么不支持,這里引用作者的說法:
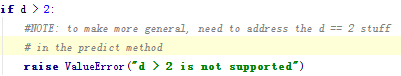
作者大概的意思是說預測方法中并沒有解決高于2階的差分,有沒有感覺很牽強,不過沒關系,我們有pandas。我們可以先用pandas將序列差分好,然后在對差分好的序列進行ARIMA擬合,只不過這樣后面會多了一步人工還原的工作。
diff_12 = ts_log.diff(12) diff_12.dropna(inplace=True) diff_12_1 = diff_12.diff(1) diff_12_1.dropna(inplace=True) test_stationarity.testStationarity(diff_12_1)

從上面的統計檢驗結果可以看出,經過12階差分和1階差分后,該序列滿足平穩性的要求了。
d. 分解
所謂分解就是將時序數據分離成不同的成分。statsmodels使用的X-11分解過程,它主要將時序數據分離成長期趨勢、季節趨勢和隨機成分。與其它統計軟件一樣,statsmodels也支持兩類分解模型,加法模型和乘法模型,這里我只實現加法,乘法只需將model的參數設置為"multiplicative"即可。
from statsmodels.tsa.seasonal import seasonal_decompose decomposition = seasonal_decompose(ts_log, model="additive") trend = decomposition.trend seasonal = decomposition.seasonal residual = decomposition.resid
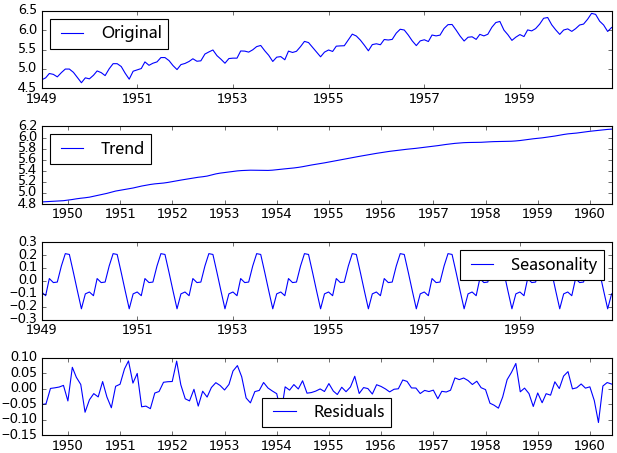
得到不同的分解成分后,就可以使用時間序列模型對各個成分進行擬合,當然也可以選擇其他預測方法。我曾經用過小波對時序數據進行過分解,然后分別采用時間序列擬合,效果還不錯。由于我對小波的理解不是很好,只能簡單的調用接口,如果有誰對小波、傅里葉、卡爾曼理解得比較透,可以將時序數據進行更加準確的分解,由于分解后的時序數據避免了他們在建模時的交叉影響,所以我相信它將有助于預測準確性的提高。
4. 模型識別
在前面的分析可知,該序列具有明顯的年周期與長期成分。對于年周期成分我們使用窗口為12的移動平進行處理,對于長期趨勢成分我們采用1階差分來進行處理。
rol_mean = ts_log.rolling(window=12).mean() rol_mean.dropna(inplace=True) ts_diff_1 = rol_mean.diff(1) ts_diff_1.dropna(inplace=True) test_stationarity.testStationarity(ts_diff_1)

觀察其統計量發現該序列在置信水平為95%的區間下并不顯著,我們對其進行再次一階差分。再次差分后的序列其自相關具有快速衰減的特點,t統計量在99%的置信水平下是顯著的,這里我不再做詳細說明。
ts_diff_2 = ts_diff_1.diff(1) ts_diff_2.dropna(inplace=True)
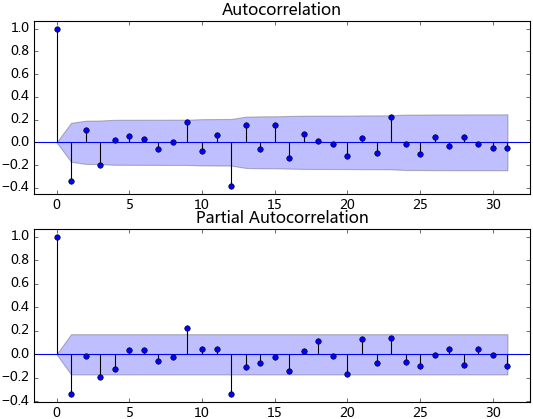
數據平穩后,需要對模型定階,即確定p、q的階數。觀察上圖,發現自相關和偏相系數都存在拖尾的特點,并且他們都具有明顯的一階相關性,所以我們設定p=1, q=1。下面就可以使用ARMA模型進行數據擬合了。這里我不使用ARIMA(ts_diff_1, order=(1, 1, 1))進行擬合,是因為含有差分操作時,預測結果還原老出問題,至今還沒弄明白。
from statsmodels.tsa.arima_model import ARMA model = ARMA(ts_diff_2, order=(1, 1)) result_arma = model.fit( disp=-1, method='css')
5. 樣本擬合
模型擬合完后,我們就可以對其進行預測了。由于ARMA擬合的是經過相關預處理后的數據,故其預測值需要通過相關逆變換進行還原。
predict_ts = result_arma.predict() # 一階差分還原diff_shift_ts = ts_diff_1.shift(1)diff_recover_1 = predict_ts.add(diff_shift_ts)# 再次一階差分還原 rol_shift_ts = rol_mean.shift(1) diff_recover = diff_recover_1.add(rol_shift_ts) # 移動平均還原 rol_sum = ts_log.rolling(window=11).sum() rol_recover = diff_recover*12 - rol_sum.shift(1) # 對數還原 log_recover = np.exp(rol_recover) log_recover.dropna(inplace=True)
我們使用均方根誤差(RMSE)來評估模型樣本內擬合的好壞。利用該準則進行判別時,需要剔除“非預測”數據的影響。
ts = ts[log_recover.index] # 過濾沒有預測的記錄plt.figure(facecolor='white')
log_recover.plot(color='blue', label='Predict')
ts.plot(color='red', label='Original')
plt.legend(loc='best')
plt.title('RMSE: %.4f'% np.sqrt(sum((log_recover-ts)**2)/ts.size))
plt.show()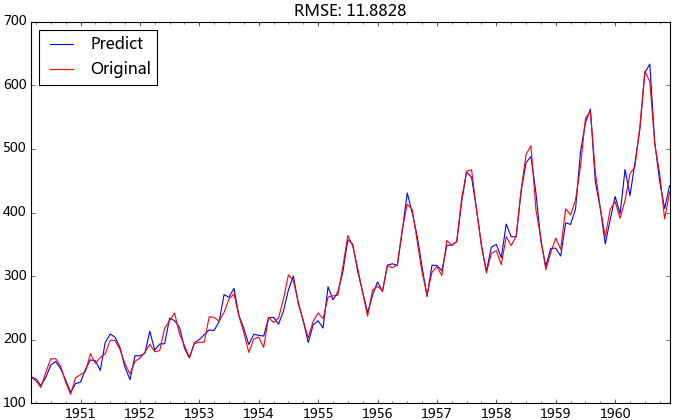
觀察上圖的擬合效果,均方根誤差為11.8828,感覺還過得去。
6.完善ARIMA模型
前面提到statsmodels里面的ARIMA模塊不支持高階差分,我們的做法是將差分分離出來,但是這樣會多了一步人工還原的操作。基于上述問題,我將差分過程進行了封裝,使序列能按照指定的差分列表依次進行差分,并相應的構造了一個還原的方法,實現差分序列的自動還原。
# 差分操作
def diff_ts(ts, d):
global shift_ts_list
# 動態預測第二日的值時所需要的差分序列
global last_data_shift_list
shift_ts_list = []
last_data_shift_list = []
tmp_ts = ts
for i in d:
last_data_shift_list.append(tmp_ts[-i])
print last_data_shift_list
shift_ts = tmp_ts.shift(i)
shift_ts_list.append(shift_ts)
tmp_ts = tmp_ts - shift_ts
tmp_ts.dropna(inplace=True)
return tmp_ts
# 還原操作
def predict_diff_recover(predict_value, d):
if isinstance(predict_value, float):
tmp_data = predict_value
for i in range(len(d)):
tmp_data = tmp_data + last_data_shift_list[-i-1]
elif isinstance(predict_value, np.ndarray):
tmp_data = predict_value[0]
for i in range(len(d)):
tmp_data = tmp_data + last_data_shift_list[-i-1]
else:
tmp_data = predict_value
for i in range(len(d)):
try:
tmp_data = tmp_data.add(shift_ts_list[-i-1])
except:
raise ValueError('What you input is not pd.Series type!')
tmp_data.dropna(inplace=True)
return tmp_data現在我們直接使用差分的方法進行數據處理,并以同樣的過程進行數據預測與還原。
diffed_ts = diff_ts(ts_log, d=[12, 1]) model = arima_model(diffed_ts) model.certain_model(1, 1) predict_ts = model.properModel.predict() diff_recover_ts = predict_diff_recover(predict_ts, d=[12, 1]) log_recover = np.exp(diff_recover_ts)

是不是發現這里的預測結果和上一篇的使用12階移動平均的預測結果一模一樣。這是因為12階移動平均加上一階差分與直接12階差分是等價的關系,后者是前者數值的12倍,這個應該不難推導。
對于個數不多的時序數據,我們可以通過觀察自相關圖和偏相關圖來進行模型識別,倘若我們要分析的時序數據量較多,例如要預測每只股票的走勢,我們就不可能逐個去調參了。這時我們可以依據BIC準則識別模型的p, q值,通常認為BIC值越小的模型相對更優。這里我簡單介紹一下BIC準則,它綜合考慮了殘差大小和自變量的個數,殘差越小BIC值越小,自變量個數越多BIC值越大。個人覺得BIC準則就是對模型過擬合設定了一個標準(過擬合這東西應該以辯證的眼光看待)。
def proper_model(data_ts, maxLag):
init_bic = sys.maxint
init_p = 0
init_q = 0
init_properModel = None
for p in np.arange(maxLag):
for q in np.arange(maxLag):
model = ARMA(data_ts, order=(p, q))
try:
results_ARMA = model.fit(disp=-1, method='css')
except:
continue
bic = results_ARMA.bic
if bic < init_bic:
init_p = p
init_q = q
init_properModel = results_ARMA
init_bic = bic
return init_bic, init_p, init_q, init_properModel相對最優參數識別結果:BIC: -1090.44209358 p: 0 q: 1 ,RMSE:11.8817198331。我們發現模型自動識別的參數要比我手動選取的參數更優。
7.滾動預測
所謂滾動預測是指通過添加最新的數據預測第二天的值。對于一個穩定的預測模型,不需要每天都去擬合,我們可以給他設定一個閥值,例如每周擬合一次,該期間只需通過添加最新的數據實現滾動預測即可。基于此我編寫了一個名為arima_model的類,主要包含模型自動識別方法,滾動預測的功能,詳細代碼可以查看附錄。數據的動態添加:
from dateutil.relativedelta import relativedeltadef _add_new_data(ts, dat, type='day'):
if type == 'day':
new_index = ts.index[-1] + relativedelta(days=1)
elif type == 'month':
new_index = ts.index[-1] + relativedelta(months=1)
ts[new_index] = dat
def add_today_data(model, ts, data, d, type='day'):
_add_new_data(ts, data, type) # 為原始序列添加數據
# 為滯后序列添加新值
d_ts = diff_ts(ts, d)
model.add_today_data(d_ts[-1], type)
def forecast_next_day_data(model, type='day'):
if model == None:
raise ValueError('No model fit before')
fc = model.forecast_next_day_value(type)
return predict_diff_recover(fc, [12, 1])現在我們就可以使用滾動預測的方法向外預測了,取1957年之前的數據作為訓練數據,其后的數據作為測試,并設定模型每第七天就會重新擬合一次。這里的diffed_ts對象會隨著add_today_data方法自動添加數據,這是由于它與add_today_data方法中的d_ts指向的同一對象,該對象會動態的添加數據。
ts_train = ts_log[:'1956-12']
ts_test = ts_log['1957-1':]
diffed_ts = diff_ts(ts_train, [12, 1])
forecast_list = []
for i, dta in enumerate(ts_test):
if i%7 == 0:
model = arima_model(diffed_ts)
model.certain_model(1, 1)
forecast_data = forecast_next_day_data(model, type='month')
forecast_list.append(forecast_data)
add_today_data(model, ts_train, dta, [12, 1], type='month')
predict_ts = pd.Series(data=forecast_list, index=ts['1957-1':].index)log_recover = np.exp(predict_ts)original_ts = ts['1957-1':]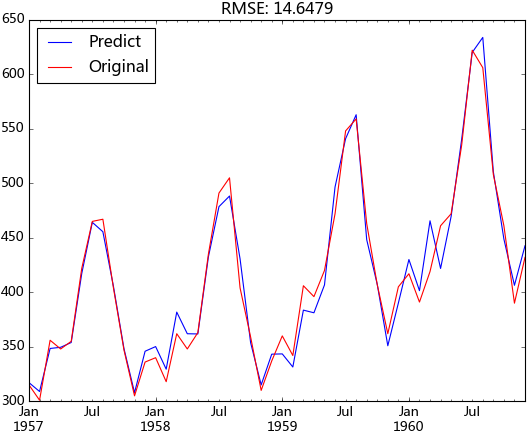
動態預測的均方根誤差為:14.6479,與前面樣本內擬合的均方根誤差相差不大,說明模型并沒有過擬合,并且整體預測效果都較好。
8. 模型序列化
在進行動態預測時,我們不希望將整個模型一直在內存中運行,而是希望有新的數據到來時才啟動該模型。這時我們就應該把整個模型從內存導出到硬盤中,而序列化正好能滿足該要求。序列化最常用的就是使用json模塊了,但是它是時間對象支持得不是很好,有人對json模塊進行了拓展以使得支持時間對象,這里我們不采用該方法,我們使用pickle模塊,它和json的接口基本相同,有興趣的可以去查看一下。我在實際應用中是采用的面向對象的編程,它的序列化主要是將類的屬性序列化即可,而在面向過程的編程中,模型序列化需要將需要序列化的對象公有化,這樣會使得對前面函數的參數改動較大,我不在詳細闡述該過程。
總結
與其它統計語言相比,python在統計分析這塊還顯得不那么“專業”。隨著numpy、pandas、scipy、sklearn、gensim、statsmodels等包的推動,我相信也祝愿python在數據分析這塊越來越好。與SAS和R相比,python的時間序列模塊還不是很成熟,我這里僅起到拋磚引玉的作用,希望各位能人志士能貢獻自己的力量,使其更加完善。實際應用中我全是面向過程來編寫的,為了闡述方便,我用面向過程重新羅列了一遍,實在感覺很不方便。原本打算分三篇來寫的,還有一部分實際應用的部分,不打算再寫了,還請大家原諒。實際應用主要是具體問題具體分析,這當中第一步就是要查詢問題,這步花的時間往往會比較多,然后再是解決問題。以我前面項目遇到的問題為例,當時遇到了以下幾個典型的問題:1.周期長度不恒定的周期成分,例如每月的1號具有周期性,但每月1號與1號之間的時間間隔是不相等的;2.含有缺失值以及含有記錄為0的情況無法進行對數變換;3.節假日的影響等等。
附錄
# -*-coding:utf-8-*-
import pandas as pd
import numpy as np
from statsmodels.tsa.arima_model import ARMA
import sys
from dateutil.relativedelta import relativedelta
from copy import deepcopy
import matplotlib.pyplot as plt
class arima_model:
def __init__(self, ts, maxLag=9):
self.data_ts = ts
self.resid_ts = None
self.predict_ts = None
self.maxLag = maxLag
self.p = maxLag
self.q = maxLag
self.properModel = None
self.bic = sys.maxint
# 計算最優ARIMA模型,將相關結果賦給相應屬性
def get_proper_model(self):
self._proper_model()
self.predict_ts = deepcopy(self.properModel.predict())
self.resid_ts = deepcopy(self.properModel.resid)
# 對于給定范圍內的p,q計算擬合得最好的arima模型,這里是對差分好的數據進行擬合,故差分恒為0
def _proper_model(self):
for p in np.arange(self.maxLag):
for q in np.arange(self.maxLag):
# print p,q,self.bic
model = ARMA(self.data_ts, order=(p, q))
try:
results_ARMA = model.fit(disp=-1, method='css')
except:
continue
bic = results_ARMA.bic
# print 'bic:',bic,'self.bic:',self.bic
if bic < self.bic:
self.p = p
self.q = q
self.properModel = results_ARMA
self.bic = bic
self.resid_ts = deepcopy(self.properModel.resid)
self.predict_ts = self.properModel.predict()
# 參數確定模型
def certain_model(self, p, q):
model = ARMA(self.data_ts, order=(p, q))
try:
self.properModel = model.fit( disp=-1, method='css')
self.p = p
self.q = q
self.bic = self.properModel.bic
self.predict_ts = self.properModel.predict()
self.resid_ts = deepcopy(self.properModel.resid)
except:
print 'You can not fit the model with this parameter p,q, ' \
'please use the get_proper_model method to get the best model'
# 預測第二日的值
def forecast_next_day_value(self, type='day'):
# 我修改了statsmodels包中arima_model的源代碼,添加了constant屬性,需要先運行forecast方法,為constant賦值
self.properModel.forecast()
if self.data_ts.index[-1] != self.resid_ts.index[-1]:
raise ValueError('''The index is different in data_ts and resid_ts, please add new data to data_ts.
If you just want to forecast the next day data without add the real next day data to data_ts,
please run the predict method which arima_model included itself''')
if not self.properModel:
raise ValueError('The arima model have not computed, please run the proper_model method before')
para = self.properModel.params
# print self.properModel.params
if self.p == 0: # It will get all the value series with setting self.data_ts[-self.p:] when p is zero
ma_value = self.resid_ts[-self.q:]
values = ma_value.reindex(index=ma_value.index[::-1])
elif self.q == 0:
ar_value = self.data_ts[-self.p:]
values = ar_value.reindex(index=ar_value.index[::-1])
else:
ar_value = self.data_ts[-self.p:]
ar_value = ar_value.reindex(index=ar_value.index[::-1])
ma_value = self.resid_ts[-self.q:]
ma_value = ma_value.reindex(index=ma_value.index[::-1])
values = ar_value.append(ma_value)
predict_value = np.dot(para[1:], values) + self.properModel.constant[0]
self._add_new_data(self.predict_ts, predict_value, type)
return predict_value
# 動態添加數據函數,針對索引是月份和日分別進行處理
def _add_new_data(self, ts, dat, type='day'):
if type == 'day':
new_index = ts.index[-1] + relativedelta(days=1)
elif type == 'month':
new_index = ts.index[-1] + relativedelta(months=1)
ts[new_index] = dat
def add_today_data(self, dat, type='day'):
self._add_new_data(self.data_ts, dat, type)
if self.data_ts.index[-1] != self.predict_ts.index[-1]:
raise ValueError('You must use the forecast_next_day_value method forecast the value of today before')
self._add_new_data(self.resid_ts, self.data_ts[-1] - self.predict_ts[-1], type)
if __name__ == '__main__':
df = pd.read_csv('AirPassengers.csv', encoding='utf-8', index_col='date')
df.index = pd.to_datetime(df.index)
ts = df['x']
# 數據預處理
ts_log = np.log(ts)
rol_mean = ts_log.rolling(window=12).mean()
rol_mean.dropna(inplace=True)
ts_diff_1 = rol_mean.diff(1)
ts_diff_1.dropna(inplace=True)
ts_diff_2 = ts_diff_1.diff(1)
ts_diff_2.dropna(inplace=True)
# 模型擬合
model = arima_model(ts_diff_2)
# 這里使用模型參數自動識別
model.get_proper_model()
print 'bic:', model.bic, 'p:', model.p, 'q:', model.q
print model.properModel.forecast()[0]
print model.forecast_next_day_value(type='month')
# 預測結果還原
predict_ts = model.properModel.predict()
diff_shift_ts = ts_diff_1.shift(1)
diff_recover_1 = predict_ts.add(diff_shift_ts)
rol_shift_ts = rol_mean.shift(1)
diff_recover = diff_recover_1.add(rol_shift_ts)
rol_sum = ts_log.rolling(window=11).sum()
rol_recover = diff_recover*12 - rol_sum.shift(1)
log_recover = np.exp(rol_recover)
log_recover.dropna(inplace=True)
# 預測結果作圖
ts = ts[log_recover.index]
plt.figure(facecolor='white')
log_recover.plot(color='blue', label='Predict')
ts.plot(color='red', label='Original')
plt.legend(loc='best')
plt.title('RMSE: %.4f'% np.sqrt(sum((log_recover-ts)**2)/ts.size))
plt.show()修改的arima_model代碼
# Note: The information criteria add 1 to the number of parameters
# whenever the model has an AR or MA term since, in principle,
# the variance could be treated as a free parameter and restricted
# This code does not allow this, but it adds consistency with other
# packages such as gretl and X12-ARIMA
from __future__ import absolute_import
from statsmodels.compat.python import string_types, range
# for 2to3 with extensions
from datetime import datetime
import numpy as np
from scipy import optimize
from scipy.stats import t, norm
from scipy.signal import lfilter
from numpy import dot, log, zeros, pi
from numpy.linalg import inv
from statsmodels.tools.decorators import (cache_readonly,
resettable_cache)
import statsmodels.tsa.base.tsa_model as tsbase
import statsmodels.base.wrapper as wrap
from statsmodels.regression.linear_model import yule_walker, GLS
from statsmodels.tsa.tsatools import (lagmat, add_trend,
_ar_transparams, _ar_invtransparams,
_ma_transparams, _ma_invtransparams,
unintegrate, unintegrate_levels)
from statsmodels.tsa.vector_ar import util
from statsmodels.tsa.ar_model import AR
from statsmodels.tsa.arima_process import arma2ma
from statsmodels.tools.numdiff import approx_hess_cs, approx_fprime_cs
from statsmodels.tsa.base.datetools import _index_date
from statsmodels.tsa.kalmanf import KalmanFilter
_armax_notes = """
Notes
-----
If exogenous variables are given, then the model that is fit is
.. math::
\\phi(L)(y_t - X_t\\beta) = \\theta(L)\epsilon_t
where :math:`\\phi` and :math:`\\theta` are polynomials in the lag
operator, :math:`L`. This is the regression model with ARMA errors,
or ARMAX model. This specification is used, whether or not the model
is fit using conditional sum of square or maximum-likelihood, using
the `method` argument in
:meth:`statsmodels.tsa.arima_model.%(Model)s.fit`. Therefore, for
now, `css` and `mle` refer to estimation methods only. This may
change for the case of the `css` model in future versions.
"""
_arma_params = """\
endog : array-like
The endogenous variable.
order : iterable
The (p,q) order of the model for the number of AR parameters,
differences, and MA parameters to use.
exog : array-like, optional
An optional arry of exogenous variables. This should *not* include a
constant or trend. You can specify this in the `fit` method."""
_arma_model = "Autoregressive Moving Average ARMA(p,q) Model"
_arima_model = "Autoregressive Integrated Moving Average ARIMA(p,d,q) Model"
_arima_params = """\
endog : array-like
The endogenous variable.
order : iterable
The (p,d,q) order of the model for the number of AR parameters,
differences, and MA parameters to use.
exog : array-like, optional
An optional arry of exogenous variables. This should *not* include a
constant or trend. You can specify this in the `fit` method."""
_predict_notes = """
Notes
-----
Use the results predict method instead.
"""
_results_notes = """
Notes
-----
It is recommended to use dates with the time-series models, as the
below will probably make clear. However, if ARIMA is used without
dates and/or `start` and `end` are given as indices, then these
indices are in terms of the *original*, undifferenced series. Ie.,
given some undifferenced observations::
1970Q1, 1
1970Q2, 1.5
1970Q3, 1.25
1970Q4, 2.25
1971Q1, 1.2
1971Q2, 4.1
1970Q1 is observation 0 in the original series. However, if we fit an
ARIMA(p,1,q) model then we lose this first observation through
differencing. Therefore, the first observation we can forecast (if
using exact MLE) is index 1. In the differenced series this is index
0, but we refer to it as 1 from the original series.
"""
_predict = """
%(Model)s model in-sample and out-of-sample prediction
Parameters
----------
%(params)s
start : int, str, or datetime
Zero-indexed observation number at which to start forecasting, ie.,
the first forecast is start. Can also be a date string to
parse or a datetime type.
end : int, str, or datetime
Zero-indexed observation number at which to end forecasting, ie.,
the first forecast is start. Can also be a date string to
parse or a datetime type. However, if the dates index does not
have a fixed frequency, end must be an integer index if you
want out of sample prediction.
exog : array-like, optional
If the model is an ARMAX and out-of-sample forecasting is
requested, exog must be given. Note that you'll need to pass
`k_ar` additional lags for any exogenous variables. E.g., if you
fit an ARMAX(2, q) model and want to predict 5 steps, you need 7
observations to do this.
dynamic : bool, optional
The `dynamic` keyword affects in-sample prediction. If dynamic
is False, then the in-sample lagged values are used for
prediction. If `dynamic` is True, then in-sample forecasts are
used in place of lagged dependent variables. The first forecasted
value is `start`.
%(extra_params)s
Returns
-------
%(returns)s
%(extra_section)s
"""
_predict_returns = """predict : array
The predicted values.
"""
_arma_predict = _predict % {"Model" : "ARMA",
"params" : """
params : array-like
The fitted parameters of the model.""",
"extra_params" : "",
"returns" : _predict_returns,
"extra_section" : _predict_notes}
_arma_results_predict = _predict % {"Model" : "ARMA", "params" : "",
"extra_params" : "",
"returns" : _predict_returns,
"extra_section" : _results_notes}
_arima_predict = _predict % {"Model" : "ARIMA",
"params" : """params : array-like
The fitted parameters of the model.""",
"extra_params" : """typ : str {'linear', 'levels'}
- 'linear' : Linear prediction in terms of the differenced
endogenous variables.
- 'levels' : Predict the levels of the original endogenous
variables.\n""", "returns" : _predict_returns,
"extra_section" : _predict_notes}
_arima_results_predict = _predict % {"Model" : "ARIMA",
"params" : "",
"extra_params" :
"""typ : str {'linear', 'levels'}
- 'linear' : Linear prediction in terms of the differenced
endogenous variables.
- 'levels' : Predict the levels of the original endogenous
variables.\n""",
"returns" : _predict_returns,
"extra_section" : _results_notes}
_arima_plot_predict_example = """ Examples
--------
>>> import statsmodels.api as sm
>>> import matplotlib.pyplot as plt
>>> import pandas as pd
>>>
>>> dta = sm.datasets.sunspots.load_pandas().data[['SUNACTIVITY']]
>>> dta.index = pd.DatetimeIndex(start='1700', end='2009', freq='A')
>>> res = sm.tsa.ARMA(dta, (3, 0)).fit()
>>> fig, ax = plt.subplots()
>>> ax = dta.ix['1950':].plot(ax=ax)
>>> fig = res.plot_predict('1990', '2012', dynamic=True, ax=ax,
... plot_insample=False)
>>> plt.show()
.. plot:: plots/arma_predict_plot.py
"""
_plot_predict = ("""
Plot forecasts
""" + '\n'.join(_predict.split('\n')[2:])) % {
"params" : "",
"extra_params" : """alpha : float, optional
The confidence intervals for the forecasts are (1 - alpha)%
plot_insample : bool, optional
Whether to plot the in-sample series. Default is True.
ax : matplotlib.Axes, optional
Existing axes to plot with.""",
"returns" : """fig : matplotlib.Figure
The plotted Figure instance""",
"extra_section" : ('\n' + _arima_plot_predict_example +
'\n' + _results_notes)
}
_arima_plot_predict = ("""
Plot forecasts
""" + '\n'.join(_predict.split('\n')[2:])) % {
"params" : "",
"extra_params" : """alpha : float, optional
The confidence intervals for the forecasts are (1 - alpha)%
plot_insample : bool, optional
Whether to plot the in-sample series. Default is True.
ax : matplotlib.Axes, optional
Existing axes to plot with.""",
"returns" : """fig : matplotlib.Figure
The plotted Figure instance""",
"extra_section" : ('\n' + _arima_plot_predict_example +
'\n' +
'\n'.join(_results_notes.split('\n')[:3]) +
("""
This is hard-coded to only allow plotting of the forecasts in levels.
""") +
'\n'.join(_results_notes.split('\n')[3:]))
}
def cumsum_n(x, n):
if n:
n -= 1
x = np.cumsum(x)
return cumsum_n(x, n)
else:
return x
def _check_arima_start(start, k_ar, k_diff, method, dynamic):
if start < 0:
raise ValueError("The start index %d of the original series "
"has been differenced away" % start)
elif (dynamic or 'mle' not in method) and start < k_ar:
raise ValueError("Start must be >= k_ar for conditional MLE "
"or dynamic forecast. Got %d" % start)
def _get_predict_out_of_sample(endog, p, q, k_trend, k_exog, start, errors,
trendparam, exparams, arparams, maparams, steps,
method, exog=None):
"""
Returns endog, resid, mu of appropriate length for out of sample
prediction.
"""
if q:
resid = np.zeros(q)
if start and 'mle' in method or (start == p and not start == 0):
resid[:q] = errors[start-q:start]
elif start:
resid[:q] = errors[start-q-p:start-p]
else:
resid[:q] = errors[-q:]
else:
resid = None
y = endog
if k_trend == 1:
# use expectation not constant
if k_exog > 0:
#TODO: technically should only hold for MLE not
# conditional model. See #274.
# ensure 2-d for conformability
if np.ndim(exog) == 1 and k_exog == 1:
# have a 1d series of observations -> 2d
exog = exog[:, None]
elif np.ndim(exog) == 1:
# should have a 1d row of exog -> 2d
if len(exog) != k_exog:
raise ValueError("1d exog given and len(exog) != k_exog")
exog = exog[None, :]
X = lagmat(np.dot(exog, exparams), p, original='in', trim='both')
mu = trendparam * (1 - arparams.sum())
# arparams were reversed in unpack for ease later
mu = mu + (np.r_[1, -arparams[::-1]] * X).sum(1)[:, None]
else:
mu = trendparam * (1 - arparams.sum())
mu = np.array([mu]*steps)
elif k_exog > 0:
X = np.dot(exog, exparams)
#NOTE: you shouldn't have to give in-sample exog!
X = lagmat(X, p, original='in', trim='both')
mu = (np.r_[1, -arparams[::-1]] * X).sum(1)[:, None]
else:
mu = np.zeros(steps)
endog = np.zeros(p + steps - 1)
if p and start:
endog[:p] = y[start-p:start]
elif p:
endog[:p] = y[-p:]
return endog, resid, mu
def _arma_predict_out_of_sample(params, steps, errors, p, q, k_trend, k_exog,
endog, exog=None, start=0, method='mle'):
(trendparam, exparams,
arparams, maparams) = _unpack_params(params, (p, q), k_trend,
k_exog, reverse=True)
# print 'params:',params
# print 'arparams:',arparams,'maparams:',maparams
endog, resid, mu = _get_predict_out_of_sample(endog, p, q, k_trend, k_exog,
start, errors, trendparam,
exparams, arparams,
maparams, steps, method,
exog)
# print 'mu[-1]:',mu[-1], 'mu[0]:',mu[0]
forecast = np.zeros(steps)
if steps == 1:
if q:
return mu[0] + np.dot(arparams, endog[:p]) + np.dot(maparams,
resid[:q]), mu[0]
else:
return mu[0] + np.dot(arparams, endog[:p]), mu[0]
if q:
i = 0 # if q == 1
else:
i = -1
for i in range(min(q, steps - 1)):
fcast = (mu[i] + np.dot(arparams, endog[i:i + p]) +
np.dot(maparams[:q - i], resid[i:i + q]))
forecast[i] = fcast
endog[i+p] = fcast
for i in range(i + 1, steps - 1):
fcast = mu[i] + np.dot(arparams, endog[i:i+p])
forecast[i] = fcast
endog[i+p] = fcast
#need to do one more without updating endog
forecast[-1] = mu[-1] + np.dot(arparams, endog[steps - 1:])
return forecast, mu[-1] #Modified by me, the former is return forecast
def _arma_predict_in_sample(start, end, endog, resid, k_ar, method):
"""
Pre- and in-sample fitting for ARMA.
"""
if 'mle' in method:
fittedvalues = endog - resid # get them all then trim
else:
fittedvalues = endog[k_ar:] - resid
fv_start = start
if 'mle' not in method:
fv_start -= k_ar # start is in terms of endog index
fv_end = min(len(fittedvalues), end + 1)
return fittedvalues[fv_start:fv_end]
def _validate(start, k_ar, k_diff, dates, method):
if isinstance(start, (string_types, datetime)):
start = _index_date(start, dates)
start -= k_diff
if 'mle' not in method and start < k_ar - k_diff:
raise ValueError("Start must be >= k_ar for conditional "
"MLE or dynamic forecast. Got %s" % start)
return start
def _unpack_params(params, order, k_trend, k_exog, reverse=False):
p, q = order
k = k_trend + k_exog
maparams = params[k+p:]
arparams = params[k:k+p]
trend = params[:k_trend]
exparams = params[k_trend:k]
if reverse:
return trend, exparams, arparams[::-1], maparams[::-1]
return trend, exparams, arparams, maparams
def _unpack_order(order):
k_ar, k_ma, k = order
k_lags = max(k_ar, k_ma+1)
return k_ar, k_ma, order, k_lags
def _make_arma_names(data, k_trend, order, exog_names):
k_ar, k_ma = order
exog_names = exog_names or []
ar_lag_names = util.make_lag_names([data.ynames], k_ar, 0)
ar_lag_names = [''.join(('ar.', i)) for i in ar_lag_names]
ma_lag_names = util.make_lag_names([data.ynames], k_ma, 0)
ma_lag_names = [''.join(('ma.', i)) for i in ma_lag_names]
trend_name = util.make_lag_names('', 0, k_trend)
exog_names = trend_name + exog_names + ar_lag_names + ma_lag_names
return exog_names
def _make_arma_exog(endog, exog, trend):
k_trend = 1 # overwritten if no constant
if exog is None and trend == 'c': # constant only
exog = np.ones((len(endog), 1))
elif exog is not None and trend == 'c': # constant plus exogenous
exog = add_trend(exog, trend='c', prepend=True)
elif exog is not None and trend == 'nc':
# make sure it's not holding constant from last run
if exog.var() == 0:
exog = None
k_trend = 0
if trend == 'nc':
k_trend = 0
return k_trend, exog
def _check_estimable(nobs, n_params):
if nobs <= n_params:
raise ValueError("Insufficient degrees of freedom to estimate")
class ARMA(tsbase.TimeSeriesModel):
__doc__ = tsbase._tsa_doc % {"model" : _arma_model,
"params" : _arma_params, "extra_params" : "",
"extra_sections" : _armax_notes %
{"Model" : "ARMA"}}
def __init__(self, endog, order, exog=None, dates=None, freq=None,
missing='none'):
super(ARMA, self).__init__(endog, exog, dates, freq, missing=missing)
exog = self.data.exog # get it after it's gone through processing
_check_estimable(len(self.endog), sum(order))
self.k_ar = k_ar = order[0]
self.k_ma = k_ma = order[1]
self.k_lags = max(k_ar, k_ma+1)
self.constant = 0 #Added by me
if exog is not None:
if exog.ndim == 1:
exog = exog[:, None]
k_exog = exog.shape[1] # number of exog. variables excl. const
else:
k_exog = 0
self.k_exog = k_exog
def _fit_start_params_hr(self, order):
"""
Get starting parameters for fit.
Parameters
----------
order : iterable
(p,q,k) - AR lags, MA lags, and number of exogenous variables
including the constant.
Returns
-------
start_params : array
A first guess at the starting parameters.
Notes
-----
If necessary, fits an AR process with the laglength selected according
to best BIC. Obtain the residuals. Then fit an ARMA(p,q) model via
OLS using these residuals for a first approximation. Uses a separate
OLS regression to find the coefficients of exogenous variables.
References
----------
Hannan, E.J. and Rissanen, J. 1982. "Recursive estimation of mixed
autoregressive-moving average order." `Biometrika`. 69.1.
"""
p, q, k = order
start_params = zeros((p+q+k))
endog = self.endog.copy() # copy because overwritten
exog = self.exog
if k != 0:
ols_params = GLS(endog, exog).fit().params
start_params[:k] = ols_params
endog -= np.dot(exog, ols_params).squeeze()
if q != 0:
if p != 0:
# make sure we don't run into small data problems in AR fit
nobs = len(endog)
maxlag = int(round(12*(nobs/100.)**(1/4.)))
if maxlag >= nobs:
maxlag = nobs - 1
armod = AR(endog).fit(ic='bic', trend='nc', maxlag=maxlag)
arcoefs_tmp = armod.params
p_tmp = armod.k_ar
# it's possible in small samples that optimal lag-order
# doesn't leave enough obs. No consistent way to fix.
if p_tmp + q >= len(endog):
raise ValueError("Proper starting parameters cannot"
" be found for this order with this "
"number of observations. Use the "
"start_params argument.")
resid = endog[p_tmp:] - np.dot(lagmat(endog, p_tmp,
trim='both'),
arcoefs_tmp)
if p < p_tmp + q:
endog_start = p_tmp + q - p
resid_start = 0
else:
endog_start = 0
resid_start = p - p_tmp - q
lag_endog = lagmat(endog, p, 'both')[endog_start:]
lag_resid = lagmat(resid, q, 'both')[resid_start:]
# stack ar lags and resids
X = np.column_stack((lag_endog, lag_resid))
coefs = GLS(endog[max(p_tmp + q, p):], X).fit().params
start_params[k:k+p+q] = coefs
else:
start_params[k+p:k+p+q] = yule_walker(endog, order=q)[0]
if q == 0 and p != 0:
arcoefs = yule_walker(endog, order=p)[0]
start_params[k:k+p] = arcoefs
# check AR coefficients
if p and not np.all(np.abs(np.roots(np.r_[1, -start_params[k:k + p]]
)) < 1):
raise ValueError("The computed initial AR coefficients are not "
"stationary\nYou should induce stationarity, "
"choose a different model order, or you can\n"
"pass your own start_params.")
# check MA coefficients
elif q and not np.all(np.abs(np.roots(np.r_[1, start_params[k + p:]]
)) < 1):
return np.zeros(len(start_params)) #modified by me
raise ValueError("The computed initial MA coefficients are not "
"invertible\nYou should induce invertibility, "
"choose a different model order, or you can\n"
"pass your own start_params.")
# check MA coefficients
# print start_params
return start_params
def _fit_start_params(self, order, method):
if method != 'css-mle': # use Hannan-Rissanen to get start params
start_params = self._fit_start_params_hr(order)
else: # use CSS to get start params
func = lambda params: -self.loglike_css(params)
#start_params = [.1]*(k_ar+k_ma+k_exog) # different one for k?
start_params = self._fit_start_params_hr(order)
if self.transparams:
start_params = self._invtransparams(start_params)
bounds = [(None,)*2]*sum(order)
mlefit = optimize.fmin_l_bfgs_b(func, start_params,
approx_grad=True, m=12,
pgtol=1e-7, factr=1e3,
bounds=bounds, iprint=-1)
start_params = self._transparams(mlefit[0])
return start_params
def score(self, params):
"""
Compute the score function at params.
Notes
-----
This is a numerical approximation.
"""
return approx_fprime_cs(params, self.loglike, args=(False,))
def hessian(self, params):
"""
Compute the Hessian at params,
Notes
-----
This is a numerical approximation.
"""
return approx_hess_cs(params, self.loglike, args=(False,))
def _transparams(self, params):
"""
Transforms params to induce stationarity/invertability.
Reference
---------
Jones(1980)
"""
k_ar, k_ma = self.k_ar, self.k_ma
k = self.k_exog + self.k_trend
newparams = np.zeros_like(params)
# just copy exogenous parameters
if k != 0:
newparams[:k] = params[:k]
# AR Coeffs
if k_ar != 0:
newparams[k:k+k_ar] = _ar_transparams(params[k:k+k_ar].copy())
# MA Coeffs
if k_ma != 0:
newparams[k+k_ar:] = _ma_transparams(params[k+k_ar:].copy())
return newparams
def _invtransparams(self, start_params):
"""
Inverse of the Jones reparameterization
"""
k_ar, k_ma = self.k_ar, self.k_ma
k = self.k_exog + self.k_trend
newparams = start_params.copy()
arcoefs = newparams[k:k+k_ar]
macoefs = newparams[k+k_ar:]
# AR coeffs
if k_ar != 0:
newparams[k:k+k_ar] = _ar_invtransparams(arcoefs)
# MA coeffs
if k_ma != 0:
newparams[k+k_ar:k+k_ar+k_ma] = _ma_invtransparams(macoefs)
return newparams
def _get_predict_start(self, start, dynamic):
# do some defaults
method = getattr(self, 'method', 'mle')
k_ar = getattr(self, 'k_ar', 0)
k_diff = getattr(self, 'k_diff', 0)
if start is None:
if 'mle' in method and not dynamic:
start = 0
else:
start = k_ar
self._set_predict_start_date(start) # else it's done in super
elif isinstance(start, int):
start = super(ARMA, self)._get_predict_start(start)
else: # should be on a date
#elif 'mle' not in method or dynamic: # should be on a date
start = _validate(start, k_ar, k_diff, self.data.dates,
method)
start = super(ARMA, self)._get_predict_start(start)
_check_arima_start(start, k_ar, k_diff, method, dynamic)
return start
def _get_predict_end(self, end, dynamic=False):
# pass through so predict works for ARIMA and ARMA
return super(ARMA, self)._get_predict_end(end)
def geterrors(self, params):
"""
Get the errors of the ARMA process.
Parameters
----------
params : array-like
The fitted ARMA parameters
order : array-like
3 item iterable, with the number of AR, MA, and exogenous
parameters, including the trend
"""
#start = self._get_predict_start(start) # will be an index of a date
#end, out_of_sample = self._get_predict_end(end)
params = np.asarray(params)
k_ar, k_ma = self.k_ar, self.k_ma
k = self.k_exog + self.k_trend
method = getattr(self, 'method', 'mle')
if 'mle' in method: # use KalmanFilter to get errors
(y, k, nobs, k_ar, k_ma, k_lags, newparams, Z_mat, m, R_mat,
T_mat, paramsdtype) = KalmanFilter._init_kalman_state(params,
self)
errors = KalmanFilter.geterrors(y, k, k_ar, k_ma, k_lags, nobs,
Z_mat, m, R_mat, T_mat,
paramsdtype)
if isinstance(errors, tuple):
errors = errors[0] # non-cython version returns a tuple
else: # use scipy.signal.lfilter
y = self.endog.copy()
k = self.k_exog + self.k_trend
if k > 0:
y -= dot(self.exog, params[:k])
k_ar = self.k_ar
k_ma = self.k_ma
(trendparams, exparams,
arparams, maparams) = _unpack_params(params, (k_ar, k_ma),
self.k_trend, self.k_exog,
reverse=False)
b, a = np.r_[1, -arparams], np.r_[1, maparams]
zi = zeros((max(k_ar, k_ma)))
for i in range(k_ar):
zi[i] = sum(-b[:i+1][::-1]*y[:i+1])
e = lfilter(b, a, y, zi=zi)
errors = e[0][k_ar:]
return errors.squeeze()
def predict(self, params, start=None, end=None, exog=None, dynamic=False):
method = getattr(self, 'method', 'mle') # don't assume fit
#params = np.asarray(params)
# will return an index of a date
start = self._get_predict_start(start, dynamic)
end, out_of_sample = self._get_predict_end(end, dynamic)
if out_of_sample and (exog is None and self.k_exog > 0):
raise ValueError("You must provide exog for ARMAX")
endog = self.endog
resid = self.geterrors(params)
k_ar = self.k_ar
if out_of_sample != 0 and self.k_exog > 0:
if self.k_exog == 1 and exog.ndim == 1:
exog = exog[:, None]
# we need the last k_ar exog for the lag-polynomial
if self.k_exog > 0 and k_ar > 0:
# need the last k_ar exog for the lag-polynomial
exog = np.vstack((self.exog[-k_ar:, self.k_trend:], exog))
if dynamic:
#TODO: now that predict does dynamic in-sample it should
# also return error estimates and confidence intervals
# but how? len(endog) is not tot_obs
out_of_sample += end - start + 1
pr, ct = _arma_predict_out_of_sample(params, out_of_sample, resid,
k_ar, self.k_ma, self.k_trend,
self.k_exog, endog, exog,
start, method)
self.constant = ct
return pr
predictedvalues = _arma_predict_in_sample(start, end, endog, resid,
k_ar, method)
if out_of_sample:
forecastvalues, ct = _arma_predict_out_of_sample(params, out_of_sample,
resid, k_ar,
self.k_ma,
self.k_trend,
self.k_exog, endog,
exog, method=method)
self.constant = ct
predictedvalues = np.r_[predictedvalues, forecastvalues]
return predictedvalues
predict.__doc__ = _arma_predict
def loglike(self, params, set_sigma2=True):
"""
Compute the log-likelihood for ARMA(p,q) model
Notes
-----
Likelihood used depends on the method set in fit
"""
method = self.method
if method in ['mle', 'css-mle']:
return self.loglike_kalman(params, set_sigma2)
elif method == 'css':
return self.loglike_css(params, set_sigma2)
else:
raise ValueError("Method %s not understood" % method)
def loglike_kalman(self, params, set_sigma2=True):
"""
Compute exact loglikelihood for ARMA(p,q) model by the Kalman Filter.
"""
return KalmanFilter.loglike(params, self, set_sigma2)
def loglike_css(self, params, set_sigma2=True):
"""
Conditional Sum of Squares likelihood function.
"""
k_ar = self.k_ar
k_ma = self.k_ma
k = self.k_exog + self.k_trend
y = self.endog.copy().astype(params.dtype)
nobs = self.nobs
# how to handle if empty?
if self.transparams:
newparams = self._transparams(params)
else:
newparams = params
if k > 0:
y -= dot(self.exog, newparams[:k])
# the order of p determines how many zeros errors to set for lfilter
b, a = np.r_[1, -newparams[k:k + k_ar]], np.r_[1, newparams[k + k_ar:]]
zi = np.zeros((max(k_ar, k_ma)), dtype=params.dtype)
for i in range(k_ar):
zi[i] = sum(-b[:i + 1][::-1] * y[:i + 1])
errors = lfilter(b, a, y, zi=zi)[0][k_ar:]
ssr = np.dot(errors, errors)
sigma2 = ssr/nobs
if set_sigma2:
self.sigma2 = sigma2
llf = -nobs/2.*(log(2*pi) + log(sigma2)) - ssr/(2*sigma2)
return llf
def fit(self, start_params=None, trend='c', method="css-mle",
transparams=True, solver='lbfgs', maxiter=50, full_output=1,
disp=5, callback=None, **kwargs):
"""
Fits ARMA(p,q) model using exact maximum likelihood via Kalman filter.
Parameters
----------
start_params : array-like, optional
Starting parameters for ARMA(p,q). If None, the default is given
by ARMA._fit_start_params. See there for more information.
transparams : bool, optional
Whehter or not to transform the parameters to ensure stationarity.
Uses the transformation suggested in Jones (1980). If False,
no checking for stationarity or invertibility is done.
method : str {'css-mle','mle','css'}
This is the loglikelihood to maximize. If "css-mle", the
conditional sum of squares likelihood is maximized and its values
are used as starting values for the computation of the exact
likelihood via the Kalman filter. If "mle", the exact likelihood
is maximized via the Kalman Filter. If "css" the conditional sum
of squares likelihood is maximized. All three methods use
`start_params` as starting parameters. See above for more
information.
trend : str {'c','nc'}
Whether to include a constant or not. 'c' includes constant,
'nc' no constant.
solver : str or None, optional
Solver to be used. The default is 'lbfgs' (limited memory
Broyden-Fletcher-Goldfarb-Shanno). Other choices are 'bfgs',
'newton' (Newton-Raphson), 'nm' (Nelder-Mead), 'cg' -
(conjugate gradient), 'ncg' (non-conjugate gradient), and
'powell'. By default, the limited memory BFGS uses m=12 to
approximate the Hessian, projected gradient tolerance of 1e-8 and
factr = 1e2. You can change these by using kwargs.
maxiter : int, optional
The maximum number of function evaluations. Default is 50.
tol : float
The convergence tolerance. Default is 1e-08.
full_output : bool, optional
If True, all output from solver will be available in
the Results object's mle_retvals attribute. Output is dependent
on the solver. See Notes for more information.
disp : bool, optional
If True, convergence information is printed. For the default
l_bfgs_b solver, disp controls the frequency of the output during
the iterations. disp < 0 means no output in this case.
callback : function, optional
Called after each iteration as callback(xk) where xk is the current
parameter vector.
kwargs
See Notes for keyword arguments that can be passed to fit.
Returns
-------
statsmodels.tsa.arima_model.ARMAResults class
See also
--------
statsmodels.base.model.LikelihoodModel.fit : for more information
on using the solvers.
ARMAResults : results class returned by fit
Notes
------
If fit by 'mle', it is assumed for the Kalman Filter that the initial
unkown state is zero, and that the inital variance is
P = dot(inv(identity(m**2)-kron(T,T)),dot(R,R.T).ravel('F')).reshape(r,
r, order = 'F')
"""
k_ar = self.k_ar
k_ma = self.k_ma
# enforce invertibility
self.transparams = transparams
endog, exog = self.endog, self.exog
k_exog = self.k_exog
self.nobs = len(endog) # this is overwritten if method is 'css'
# (re)set trend and handle exogenous variables
# always pass original exog
k_trend, exog = _make_arma_exog(endog, self.exog, trend)
# Check has something to estimate
if k_ar == 0 and k_ma == 0 and k_trend == 0 and k_exog == 0:
raise ValueError("Estimation requires the inclusion of least one "
"AR term, MA term, a constant or an exogenous "
"variable.")
# check again now that we know the trend
_check_estimable(len(endog), k_ar + k_ma + k_exog + k_trend)
self.k_trend = k_trend
self.exog = exog # overwrites original exog from __init__
# (re)set names for this model
self.exog_names = _make_arma_names(self.data, k_trend, (k_ar, k_ma),
self.exog_names)
k = k_trend + k_exog
# choose objective function
if k_ma == 0 and k_ar == 0:
method = "css" # Always CSS when no AR or MA terms
self.method = method = method.lower()
# adjust nobs for css
if method == 'css':
self.nobs = len(self.endog) - k_ar
if start_params is not None:
start_params = np.asarray(start_params)
else: # estimate starting parameters
start_params = self._fit_start_params((k_ar, k_ma, k), method)
if transparams: # transform initial parameters to ensure invertibility
start_params = self._invtransparams(start_params)
if solver == 'lbfgs':
kwargs.setdefault('pgtol', 1e-8)
kwargs.setdefault('factr', 1e2)
kwargs.setdefault('m', 12)
kwargs.setdefault('approx_grad', True)
mlefit = super(ARMA, self).fit(start_params, method=solver,
maxiter=maxiter,
full_output=full_output, disp=disp,
callback=callback, **kwargs)
params = mlefit.params
if transparams: # transform parameters back
params = self._transparams(params)
self.transparams = False # so methods don't expect transf.
normalized_cov_params = None # TODO: fix this
armafit = ARMAResults(self, params, normalized_cov_params)
armafit.mle_retvals = mlefit.mle_retvals
armafit.mle_settings = mlefit.mle_settings
armafit.mlefit = mlefit
return ARMAResultsWrapper(armafit)
#NOTE: the length of endog changes when we give a difference to fit
#so model methods are not the same on unfit models as fit ones
#starting to think that order of model should be put in instantiation...
class ARIMA(ARMA):
__doc__ = tsbase._tsa_doc % {"model" : _arima_model,
"params" : _arima_params, "extra_params" : "",
"extra_sections" : _armax_notes %
{"Model" : "ARIMA"}}
def __new__(cls, endog, order, exog=None, dates=None, freq=None,
missing='none'):
p, d, q = order
if d == 0: # then we just use an ARMA model
return ARMA(endog, (p, q), exog, dates, freq, missing)
else:
mod = super(ARIMA, cls).__new__(cls)
mod.__init__(endog, order, exog, dates, freq, missing)
return mod
def __init__(self, endog, order, exog=None, dates=None, freq=None,
missing='none'):
p, d, q = order
if d > 2:
#NOTE: to make more general, need to address the d == 2 stuff
# in the predict method
raise ValueError("d > 2 is not supported")
super(ARIMA, self).__init__(endog, (p, q), exog, dates, freq, missing)
self.k_diff = d
self._first_unintegrate = unintegrate_levels(self.endog[:d], d)
self.endog = np.diff(self.endog, n=d)
#NOTE: will check in ARMA but check again since differenced now
_check_estimable(len(self.endog), p+q)
if exog is not None:
self.exog = self.exog[d:]
if d == 1:
self.data.ynames = 'D.' + self.endog_names
else:
self.data.ynames = 'D{0:d}.'.format(d) + self.endog_names
# what about exog, should we difference it automatically before
# super call?
def _get_predict_start(self, start, dynamic):
"""
"""
#TODO: remove all these getattr and move order specification to
# class constructor
k_diff = getattr(self, 'k_diff', 0)
method = getattr(self, 'method', 'mle')
k_ar = getattr(self, 'k_ar', 0)
if start is None:
if 'mle' in method and not dynamic:
start = 0
else:
start = k_ar
elif isinstance(start, int):
start -= k_diff
try: # catch when given an integer outside of dates index
start = super(ARIMA, self)._get_predict_start(start,
dynamic)
except IndexError:
raise ValueError("start must be in series. "
"got %d" % (start + k_diff))
else: # received a date
start = _validate(start, k_ar, k_diff, self.data.dates,
method)
start = super(ARIMA, self)._get_predict_start(start, dynamic)
# reset date for k_diff adjustment
self._set_predict_start_date(start + k_diff)
return start
def _get_predict_end(self, end, dynamic=False):
"""
Returns last index to be forecast of the differenced array.
Handling of inclusiveness should be done in the predict function.
"""
end, out_of_sample = super(ARIMA, self)._get_predict_end(end, dynamic)
if 'mle' not in self.method and not dynamic:
end -= self.k_ar
return end - self.k_diff, out_of_sample
def fit(self, start_params=None, trend='c', method="css-mle",
transparams=True, solver='lbfgs', maxiter=50, full_output=1,
disp=5, callback=None, **kwargs):
"""
Fits ARIMA(p,d,q) model by exact maximum likelihood via Kalman filter.
Parameters
----------
start_params : array-like, optional
Starting parameters for ARMA(p,q). If None, the default is given
by ARMA._fit_start_params. See there for more information.
transparams : bool, optional
Whehter or not to transform the parameters to ensure stationarity.
Uses the transformation suggested in Jones (1980). If False,
no checking for stationarity or invertibility is done.
method : str {'css-mle','mle','css'}
This is the loglikelihood to maximize. If "css-mle", the
conditional sum of squares likelihood is maximized and its values
are used as starting values for the computation of the exact
likelihood via the Kalman filter. If "mle", the exact likelihood
is maximized via the Kalman Filter. If "css" the conditional sum
of squares likelihood is maximized. All three methods use
`start_params` as starting parameters. See above for more
information.
trend : str {'c','nc'}
Whether to include a constant or not. 'c' includes constant,
'nc' no constant.
solver : str or None, optional
Solver to be used. The default is 'lbfgs' (limited memory
Broyden-Fletcher-Goldfarb-Shanno). Other choices are 'bfgs',
'newton' (Newton-Raphson), 'nm' (Nelder-Mead), 'cg' -
(conjugate gradient), 'ncg' (non-conjugate gradient), and
'powell'. By default, the limited memory BFGS uses m=12 to
approximate the Hessian, projected gradient tolerance of 1e-8 and
factr = 1e2. You can change these by using kwargs.
maxiter : int, optional
The maximum number of function evaluations. Default is 50.
tol : float
The convergence tolerance. Default is 1e-08.
full_output : bool, optional
If True, all output from solver will be available in
the Results object's mle_retvals attribute. Output is dependent
on the solver. See Notes for more information.
disp : bool, optional
If True, convergence information is printed. For the default
l_bfgs_b solver, disp controls the frequency of the output during
the iterations. disp < 0 means no output in this case.
callback : function, optional
Called after each iteration as callback(xk) where xk is the current
parameter vector.
kwargs
See Notes for keyword arguments that can be passed to fit.
Returns
-------
`statsmodels.tsa.arima.ARIMAResults` class
See also
--------
statsmodels.base.model.LikelihoodModel.fit : for more information
on using the solvers.
ARIMAResults : results class returned by fit
Notes
------
If fit by 'mle', it is assumed for the Kalman Filter that the initial
unkown state is zero, and that the inital variance is
P = dot(inv(identity(m**2)-kron(T,T)),dot(R,R.T).ravel('F')).reshape(r,
r, order = 'F')
"""
arima_fit = super(ARIMA, self).fit(start_params, trend,
method, transparams, solver,
maxiter, full_output, disp,
callback, **kwargs)
normalized_cov_params = None # TODO: fix this?
arima_fit = ARIMAResults(self, arima_fit._results.params,
normalized_cov_params)
arima_fit.k_diff = self.k_diff
return ARIMAResultsWrapper(arima_fit)
def predict(self, params, start=None, end=None, exog=None, typ='linear',
dynamic=False):
# go ahead and convert to an index for easier checking
if isinstance(start, (string_types, datetime)):
start = _index_date(start, self.data.dates)
if typ == 'linear':
if not dynamic or (start != self.k_ar + self.k_diff and
start is not None):
return super(ARIMA, self).predict(params, start, end, exog,
dynamic)
else:
# need to assume pre-sample residuals are zero
# do this by a hack
q = self.k_ma
self.k_ma = 0
predictedvalues = super(ARIMA, self).predict(params, start,
end, exog,
dynamic)
self.k_ma = q
return predictedvalues
elif typ == 'levels':
endog = self.data.endog
if not dynamic:
predict = super(ARIMA, self).predict(params, start, end,
dynamic)
start = self._get_predict_start(start, dynamic)
end, out_of_sample = self._get_predict_end(end)
d = self.k_diff
if 'mle' in self.method:
start += d - 1 # for case where d == 2
end += d - 1
# add each predicted diff to lagged endog
if out_of_sample:
fv = predict[:-out_of_sample] + endog[start:end+1]
if d == 2: #TODO: make a general solution to this
fv += np.diff(endog[start - 1:end + 1])
levels = unintegrate_levels(endog[-d:], d)
fv = np.r_[fv,
unintegrate(predict[-out_of_sample:],
levels)[d:]]
else:
fv = predict + endog[start:end + 1]
if d == 2:
fv += np.diff(endog[start - 1:end + 1])
else:
k_ar = self.k_ar
if out_of_sample:
fv = (predict[:-out_of_sample] +
endog[max(start, self.k_ar-1):end+k_ar+1])
if d == 2:
fv += np.diff(endog[start - 1:end + 1])
levels = unintegrate_levels(endog[-d:], d)
fv = np.r_[fv,
unintegrate(predict[-out_of_sample:],
levels)[d:]]
else:
fv = predict + endog[max(start, k_ar):end+k_ar+1]
if d == 2:
fv += np.diff(endog[start - 1:end + 1])
else:
#IFF we need to use pre-sample values assume pre-sample
# residuals are zero, do this by a hack
if start == self.k_ar + self.k_diff or start is None:
# do the first k_diff+1 separately
p = self.k_ar
q = self.k_ma
k_exog = self.k_exog
k_trend = self.k_trend
k_diff = self.k_diff
(trendparam, exparams,
arparams, maparams) = _unpack_params(params, (p, q),
k_trend,
k_exog,
reverse=True)
# this is the hack
self.k_ma = 0
predict = super(ARIMA, self).predict(params, start, end,
exog, dynamic)
if not start:
start = self._get_predict_start(start, dynamic)
start += k_diff
self.k_ma = q
return endog[start-1] + np.cumsum(predict)
else:
predict = super(ARIMA, self).predict(params, start, end,
exog, dynamic)
return endog[start-1] + np.cumsum(predict)
return fv
else: # pragma : no cover
raise ValueError("typ %s not understood" % typ)
predict.__doc__ = _arima_predict
class ARMAResults(tsbase.TimeSeriesModelResults):
"""
Class to hold results from fitting an ARMA model.
Parameters
----------
model : ARMA instance
The fitted model instance
params : array
Fitted parameters
normalized_cov_params : array, optional
The normalized variance covariance matrix
scale : float, optional
Optional argument to scale the variance covariance matrix.
Returns
--------
**Attributes**
aic : float
Akaike Information Criterion
:math:`-2*llf+2* df_model`
where `df_model` includes all AR parameters, MA parameters, constant
terms parameters on constant terms and the variance.
arparams : array
The parameters associated with the AR coefficients in the model.
arroots : array
The roots of the AR coefficients are the solution to
(1 - arparams[0]*z - arparams[1]*z**2 -...- arparams[p-1]*z**k_ar) = 0
Stability requires that the roots in modulus lie outside the unit
circle.
bic : float
Bayes Information Criterion
-2*llf + log(nobs)*df_model
Where if the model is fit using conditional sum of squares, the
number of observations `nobs` does not include the `p` pre-sample
observations.
bse : array
The standard errors of the parameters. These are computed using the
numerical Hessian.
df_model : array
The model degrees of freedom = `k_exog` + `k_trend` + `k_ar` + `k_ma`
df_resid : array
The residual degrees of freedom = `nobs` - `df_model`
fittedvalues : array
The predicted values of the model.
hqic : float
Hannan-Quinn Information Criterion
-2*llf + 2*(`df_model`)*log(log(nobs))
Like `bic` if the model is fit using conditional sum of squares then
the `k_ar` pre-sample observations are not counted in `nobs`.
k_ar : int
The number of AR coefficients in the model.
k_exog : int
The number of exogenous variables included in the model. Does not
include the constant.
k_ma : int
The number of MA coefficients.
k_trend : int
This is 0 for no constant or 1 if a constant is included.
llf : float
The value of the log-likelihood function evaluated at `params`.
maparams : array
The value of the moving average coefficients.
maroots : array
The roots of the MA coefficients are the solution to
(1 + maparams[0]*z + maparams[1]*z**2 + ... + maparams[q-1]*z**q) = 0
Stability requires that the roots in modules lie outside the unit
circle.
model : ARMA instance
A reference to the model that was fit.
nobs : float
The number of observations used to fit the model. If the model is fit
using exact maximum likelihood this is equal to the total number of
observations, `n_totobs`. If the model is fit using conditional
maximum likelihood this is equal to `n_totobs` - `k_ar`.
n_totobs : float
The total number of observations for `endog`. This includes all
observations, even pre-sample values if the model is fit using `css`.
params : array
The parameters of the model. The order of variables is the trend
coefficients and the `k_exog` exognous coefficients, then the
`k_ar` AR coefficients, and finally the `k_ma` MA coefficients.
pvalues : array
The p-values associated with the t-values of the coefficients. Note
that the coefficients are assumed to have a Student's T distribution.
resid : array
The model residuals. If the model is fit using 'mle' then the
residuals are created via the Kalman Filter. If the model is fit
using 'css' then the residuals are obtained via `scipy.signal.lfilter`
adjusted such that the first `k_ma` residuals are zero. These zero
residuals are not returned.
scale : float
This is currently set to 1.0 and not used by the model or its results.
sigma2 : float
The variance of the residuals. If the model is fit by 'css',
sigma2 = ssr/nobs, where ssr is the sum of squared residuals. If
the model is fit by 'mle', then sigma2 = 1/nobs * sum(v**2 / F)
where v is the one-step forecast error and F is the forecast error
variance. See `nobs` for the difference in definitions depending on the
fit.
"""
_cache = {}
#TODO: use this for docstring when we fix nobs issue
def __init__(self, model, params, normalized_cov_params=None, scale=1.):
super(ARMAResults, self).__init__(model, params, normalized_cov_params,
scale)
self.sigma2 = model.sigma2
nobs = model.nobs
self.nobs = nobs
k_exog = model.k_exog
self.k_exog = k_exog
k_trend = model.k_trend
self.k_trend = k_trend
k_ar = model.k_ar
self.k_ar = k_ar
self.n_totobs = len(model.endog)
k_ma = model.k_ma
self.k_ma = k_ma
df_model = k_exog + k_trend + k_ar + k_ma
self._ic_df_model = df_model + 1
self.df_model = df_model
self.df_resid = self.nobs - df_model
self._cache = resettable_cache()
self.constant = 0 #Added by me
@cache_readonly
def arroots(self):
return np.roots(np.r_[1, -self.arparams])**-1
@cache_readonly
def maroots(self):
return np.roots(np.r_[1, self.maparams])**-1
@cache_readonly
def arfreq(self):
r"""
Returns the frequency of the AR roots.
This is the solution, x, to z = abs(z)*exp(2j*np.pi*x) where z are the
roots.
"""
z = self.arroots
if not z.size:
return
return np.arctan2(z.imag, z.real) / (2*pi)
@cache_readonly
def mafreq(self):
r"""
Returns the frequency of the MA roots.
This is the solution, x, to z = abs(z)*exp(2j*np.pi*x) where z are the
roots.
"""
z = self.maroots
if not z.size:
return
return np.arctan2(z.imag, z.real) / (2*pi)
@cache_readonly
def arparams(self):
k = self.k_exog + self.k_trend
return self.params[k:k+self.k_ar]
@cache_readonly
def maparams(self):
k = self.k_exog + self.k_trend
k_ar = self.k_ar
return self.params[k+k_ar:]
@cache_readonly
def llf(self):
return self.model.loglike(self.params)
@cache_readonly
def bse(self):
params = self.params
hess = self.model.hessian(params)
if len(params) == 1: # can't take an inverse, ensure 1d
return np.sqrt(-1./hess[0])
return np.sqrt(np.diag(-inv(hess)))
def cov_params(self): # add scale argument?
params = self.params
hess = self.model.hessian(params)
return -inv(hess)
@cache_readonly
def aic(self):
return -2 * self.llf + 2 * self._ic_df_model
@cache_readonly
def bic(self):
nobs = self.nobs
return -2 * self.llf + np.log(nobs) * self._ic_df_model
@cache_readonly
def hqic(self):
nobs = self.nobs
return -2 * self.llf + 2 * np.log(np.log(nobs)) * self._ic_df_model
@cache_readonly
def fittedvalues(self):
model = self.model
endog = model.endog.copy()
k_ar = self.k_ar
exog = model.exog # this is a copy
if exog is not None:
if model.method == "css" and k_ar > 0:
exog = exog[k_ar:]
if model.method == "css" and k_ar > 0:
endog = endog[k_ar:]
fv = endog - self.resid
# add deterministic part back in
#k = self.k_exog + self.k_trend
#TODO: this needs to be commented out for MLE with constant
#if k != 0:
# fv += dot(exog, self.params[:k])
return fv
@cache_readonly
def resid(self):
return self.model.geterrors(self.params)
@cache_readonly
def pvalues(self):
#TODO: same for conditional and unconditional?
df_resid = self.df_resid
return t.sf(np.abs(self.tvalues), df_resid) * 2
def predict(self, start=None, end=None, exog=None, dynamic=False):
return self.model.predict(self.params, start, end, exog, dynamic)
predict.__doc__ = _arma_results_predict
def _forecast_error(self, steps):
sigma2 = self.sigma2
ma_rep = arma2ma(np.r_[1, -self.arparams],
np.r_[1, self.maparams], nobs=steps)
fcasterr = np.sqrt(sigma2 * np.cumsum(ma_rep**2))
return fcasterr
def _forecast_conf_int(self, forecast, fcasterr, alpha):
const = norm.ppf(1 - alpha / 2.)
conf_int = np.c_[forecast - const * fcasterr,
forecast + const * fcasterr]
return conf_int
def forecast(self, steps=1, exog=None, alpha=.05):
"""
Out-of-sample forecasts
Parameters
----------
steps : int
The number of out of sample forecasts from the end of the
sample.
exog : array
If the model is an ARMAX, you must provide out of sample
values for the exogenous variables. This should not include
the constant.
alpha : float
The confidence intervals for the forecasts are (1 - alpha) %
Returns
-------
forecast : array
Array of out of sample forecasts
stderr : array
Array of the standard error of the forecasts.
conf_int : array
2d array of the confidence interval for the forecast
"""
if exog is not None:
#TODO: make a convenience function for this. we're using the
# pattern elsewhere in the codebase
exog = np.asarray(exog)
if self.k_exog == 1 and exog.ndim == 1:
exog = exog[:, None]
elif exog.ndim == 1:
if len(exog) != self.k_exog:
raise ValueError("1d exog given and len(exog) != k_exog")
exog = exog[None, :]
if exog.shape[0] != steps:
raise ValueError("new exog needed for each step")
# prepend in-sample exog observations
exog = np.vstack((self.model.exog[-self.k_ar:, self.k_trend:],
exog))
forecast, ct = _arma_predict_out_of_sample(self.params,
steps, self.resid, self.k_ar,
self.k_ma, self.k_trend,
self.k_exog, self.model.endog,
exog, method=self.model.method)
self.constant = ct
# compute the standard errors
fcasterr = self._forecast_error(steps)
conf_int = self._forecast_conf_int(forecast, fcasterr, alpha)
return forecast, fcasterr, conf_int
def summary(self, alpha=.05):
"""Summarize the Model
Parameters
----------
alpha : float, optional
Significance level for the confidence intervals.
Returns
-------
smry : Summary instance
This holds the summary table and text, which can be printed or
converted to various output formats.
See Also
--------
statsmodels.iolib.summary.Summary
"""
from statsmodels.iolib.summary import Summary
model = self.model
title = model.__class__.__name__ + ' Model Results'
method = model.method
# get sample TODO: make better sample machinery for estimation
k_diff = getattr(self, 'k_diff', 0)
if 'mle' in method:
start = k_diff
else:
start = k_diff + self.k_ar
if self.data.dates is not None:
dates = self.data.dates
sample = [dates[start].strftime('%m-%d-%Y')]
sample += ['- ' + dates[-1].strftime('%m-%d-%Y')]
else:
sample = str(start) + ' - ' + str(len(self.data.orig_endog))
k_ar, k_ma = self.k_ar, self.k_ma
if not k_diff:
order = str((k_ar, k_ma))
else:
order = str((k_ar, k_diff, k_ma))
top_left = [('Dep. Variable:', None),
('Model:', [model.__class__.__name__ + order]),
('Method:', [method]),
('Date:', None),
('Time:', None),
('Sample:', [sample[0]]),
('', [sample[1]])
]
top_right = [
('No. Observations:', [str(len(self.model.endog))]),
('Log Likelihood', ["%#5.3f" % self.llf]),
('S.D. of innovations', ["%#5.3f" % self.sigma2**.5]),
('AIC', ["%#5.3f" % self.aic]),
('BIC', ["%#5.3f" % self.bic]),
('HQIC', ["%#5.3f" % self.hqic])]
smry = Summary()
smry.add_table_2cols(self, gleft=top_left, gright=top_right,
title=title)
smry.add_table_params(self, alpha=alpha, use_t=False)
# Make the roots table
from statsmodels.iolib.table import SimpleTable
if k_ma and k_ar:
arstubs = ["AR.%d" % i for i in range(1, k_ar + 1)]
mastubs = ["MA.%d" % i for i in range(1, k_ma + 1)]
stubs = arstubs + mastubs
roots = np.r_[self.arroots, self.maroots]
freq = np.r_[self.arfreq, self.mafreq]
elif k_ma:
mastubs = ["MA.%d" % i for i in range(1, k_ma + 1)]
stubs = mastubs
roots = self.maroots
freq = self.mafreq
elif k_ar:
arstubs = ["AR.%d" % i for i in range(1, k_ar + 1)]
stubs = arstubs
roots = self.arroots
freq = self.arfreq
else: # 0,0 model
stubs = []
if len(stubs): # not 0, 0
modulus = np.abs(roots)
data = np.column_stack((roots.real, roots.imag, modulus, freq))
roots_table = SimpleTable(data,
headers=[' Real',
' Imaginary',
' Modulus',
' Frequency'],
title="Roots",
stubs=stubs,
data_fmts=["%17.4f", "%+17.4fj",
"%17.4f", "%17.4f"])
smry.tables.append(roots_table)
return smry
def summary2(self, title=None, alpha=.05, float_format="%.4f"):
"""Experimental summary function for ARIMA Results
Parameters
-----------
title : string, optional
Title for the top table. If not None, then this replaces the
default title
alpha : float
significance level for the confidence intervals
float_format: string
print format for floats in parameters summary
Returns
-------
smry : Summary instance
This holds the summary table and text, which can be printed or
converted to various output formats.
See Also
--------
statsmodels.iolib.summary2.Summary : class to hold summary
results
"""
from pandas import DataFrame
# get sample TODO: make better sample machinery for estimation
k_diff = getattr(self, 'k_diff', 0)
if 'mle' in self.model.method:
start = k_diff
else:
start = k_diff + self.k_ar
if self.data.dates is not None:
dates = self.data.dates
sample = [dates[start].strftime('%m-%d-%Y')]
sample += [dates[-1].strftime('%m-%d-%Y')]
else:
sample = str(start) + ' - ' + str(len(self.data.orig_endog))
k_ar, k_ma = self.k_ar, self.k_ma
# Roots table
if k_ma and k_ar:
arstubs = ["AR.%d" % i for i in range(1, k_ar + 1)]
mastubs = ["MA.%d" % i for i in range(1, k_ma + 1)]
stubs = arstubs + mastubs
roots = np.r_[self.arroots, self.maroots]
freq = np.r_[self.arfreq, self.mafreq]
elif k_ma:
mastubs = ["MA.%d" % i for i in range(1, k_ma + 1)]
stubs = mastubs
roots = self.maroots
freq = self.mafreq
elif k_ar:
arstubs = ["AR.%d" % i for i in range(1, k_ar + 1)]
stubs = arstubs
roots = self.arroots
freq = self.arfreq
else: # 0, 0 order
stubs = []
if len(stubs):
modulus = np.abs(roots)
data = np.column_stack((roots.real, roots.imag, modulus, freq))
data = DataFrame(data)
data.columns = ['Real', 'Imaginary', 'Modulus', 'Frequency']
data.index = stubs
# Summary
from statsmodels.iolib import summary2
smry = summary2.Summary()
# Model info
model_info = summary2.summary_model(self)
model_info['Method:'] = self.model.method
model_info['Sample:'] = sample[0]
model_info[' '] = sample[-1]
model_info['S.D. of innovations:'] = "%#5.3f" % self.sigma2**.5
model_info['HQIC:'] = "%#5.3f" % self.hqic
model_info['No. Observations:'] = str(len(self.model.endog))
# Parameters
params = summary2.summary_params(self)
smry.add_dict(model_info)
smry.add_df(params, float_format=float_format)
if len(stubs):
smry.add_df(data, float_format="%17.4f")
smry.add_title(results=self, title=title)
return smry
def plot_predict(self, start=None, end=None, exog=None, dynamic=False,
alpha=.05, plot_insample=True, ax=None):
from statsmodels.graphics.utils import _import_mpl, create_mpl_ax
_ = _import_mpl()
fig, ax = create_mpl_ax(ax)
# use predict so you set dates
forecast = self.predict(start, end, exog, dynamic)
# doing this twice. just add a plot keyword to predict?
start = self.model._get_predict_start(start, dynamic=False)
end, out_of_sample = self.model._get_predict_end(end, dynamic=False)
if out_of_sample:
steps = out_of_sample
fc_error = self._forecast_error(steps)
conf_int = self._forecast_conf_int(forecast[-steps:], fc_error,
alpha)
if hasattr(self.data, "predict_dates"):
from pandas import TimeSeries
forecast = TimeSeries(forecast, index=self.data.predict_dates)
ax = forecast.plot(ax=ax, label='forecast')
else:
ax.plot(forecast)
x = ax.get_lines()[-1].get_xdata()
if out_of_sample:
label = "{0:.0%} confidence interval".format(1 - alpha)
ax.fill_between(x[-out_of_sample:], conf_int[:, 0], conf_int[:, 1],
color='gray', alpha=.5, label=label)
if plot_insample:
ax.plot(x[:end + 1 - start], self.model.endog[start:end+1],
label=self.model.endog_names)
ax.legend(loc='best')
return fig
plot_predict.__doc__ = _plot_predict
class ARMAResultsWrapper(wrap.ResultsWrapper):
_attrs = {}
_wrap_attrs = wrap.union_dicts(tsbase.TimeSeriesResultsWrapper._wrap_attrs,
_attrs)
_methods = {}
_wrap_methods = wrap.union_dicts(tsbase.TimeSeriesResultsWrapper._wrap_methods,
_methods)
wrap.populate_wrapper(ARMAResultsWrapper, ARMAResults)
class ARIMAResults(ARMAResults):
def predict(self, start=None, end=None, exog=None, typ='linear',
dynamic=False):
return self.model.predict(self.params, start, end, exog, typ, dynamic)
predict.__doc__ = _arima_results_predict
def _forecast_error(self, steps):
sigma2 = self.sigma2
ma_rep = arma2ma(np.r_[1, -self.arparams],
np.r_[1, self.maparams], nobs=steps)
fcerr = np.sqrt(np.cumsum(cumsum_n(ma_rep, self.k_diff)**2)*sigma2)
return fcerr
def _forecast_conf_int(self, forecast, fcerr, alpha):
const = norm.ppf(1 - alpha/2.)
conf_int = np.c_[forecast - const*fcerr, forecast + const*fcerr]
return conf_int
def forecast(self, steps=1, exog=None, alpha=.05):
"""
Out-of-sample forecasts
Parameters
----------
steps : int
The number of out of sample forecasts from the end of the
sample.
exog : array
If the model is an ARIMAX, you must provide out of sample
values for the exogenous variables. This should not include
the constant.
alpha : float
The confidence intervals for the forecasts are (1 - alpha) %
Returns
-------
forecast : array
Array of out of sample forecasts
stderr : array
Array of the standard error of the forecasts.
conf_int : array
2d array of the confidence interval for the forecast
Notes
-----
Prediction is done in the levels of the original endogenous variable.
If you would like prediction of differences in levels use `predict`.
"""
if exog is not None:
if self.k_exog == 1 and exog.ndim == 1:
exog = exog[:, None]
if exog.shape[0] != steps:
raise ValueError("new exog needed for each step")
# prepend in-sample exog observations
exog = np.vstack((self.model.exog[-self.k_ar:, self.k_trend:],
exog))
forecast, ct = _arma_predict_out_of_sample(self.params, steps, self.resid,
self.k_ar, self.k_ma,
self.k_trend, self.k_exog,
self.model.endog,
exog, method=self.model.method)
#self.constant = ct
d = self.k_diff
endog = self.model.data.endog[-d:]
forecast = unintegrate(forecast, unintegrate_levels(endog, d))[d:]
# get forecast errors
fcerr = self._forecast_error(steps)
conf_int = self._forecast_conf_int(forecast, fcerr, alpha)
return forecast, fcerr, conf_int
def plot_predict(self, start=None, end=None, exog=None, dynamic=False,
alpha=.05, plot_insample=True, ax=None):
from statsmodels.graphics.utils import _import_mpl, create_mpl_ax
_ = _import_mpl()
fig, ax = create_mpl_ax(ax)
# use predict so you set dates
forecast = self.predict(start, end, exog, 'levels', dynamic)
# doing this twice. just add a plot keyword to predict?
start = self.model._get_predict_start(start, dynamic=dynamic)
end, out_of_sample = self.model._get_predict_end(end, dynamic=dynamic)
if out_of_sample:
steps = out_of_sample
fc_error = self._forecast_error(steps)
conf_int = self._forecast_conf_int(forecast[-steps:], fc_error,
alpha)
if hasattr(self.data, "predict_dates"):
from pandas import TimeSeries
forecast = TimeSeries(forecast, index=self.data.predict_dates)
ax = forecast.plot(ax=ax, label='forecast')
else:
ax.plot(forecast)
x = ax.get_lines()[-1].get_xdata()
if out_of_sample:
label = "{0:.0%} confidence interval".format(1 - alpha)
ax.fill_between(x[-out_of_sample:], conf_int[:, 0], conf_int[:, 1],
color='gray', alpha=.5, label=label)
if plot_insample:
import re
k_diff = self.k_diff
label = re.sub("D\d*\.", "", self.model.endog_names)
levels = unintegrate(self.model.endog,
self.model._first_unintegrate)
ax.plot(x[:end + 1 - start],
levels[start + k_diff:end + k_diff + 1], label=label)
ax.legend(loc='best')
return fig
plot_predict.__doc__ = _arima_plot_predict
class ARIMAResultsWrapper(ARMAResultsWrapper):
pass
wrap.populate_wrapper(ARIMAResultsWrapper, ARIMAResults)
if __name__ == "__main__":
import statsmodels.api as sm
# simulate arma process
from statsmodels.tsa.arima_process import arma_generate_sample
y = arma_generate_sample([1., -.75], [1., .25], nsample=1000)
arma = ARMA(y)
res = arma.fit(trend='nc', order=(1, 1))
np.random.seed(12345)
y_arma22 = arma_generate_sample([1., -.85, .35], [1, .25, -.9],
nsample=1000)
arma22 = ARMA(y_arma22)
res22 = arma22.fit(trend='nc', order=(2, 2))
# test CSS
arma22_css = ARMA(y_arma22)
res22css = arma22_css.fit(trend='nc', order=(2, 2), method='css')
data = sm.datasets.sunspots.load()
ar = ARMA(data.endog)
resar = ar.fit(trend='nc', order=(9, 0))
y_arma31 = arma_generate_sample([1, -.75, -.35, .25], [.1],
nsample=1000)
arma31css = ARMA(y_arma31)
res31css = arma31css.fit(order=(3, 1), method="css", trend="nc",
transparams=True)
y_arma13 = arma_generate_sample([1., -.75], [1, .25, -.5, .8],
nsample=1000)
arma13css = ARMA(y_arma13)
res13css = arma13css.fit(order=(1, 3), method='css', trend='nc')
# check css for p < q and q < p
y_arma41 = arma_generate_sample([1., -.75, .35, .25, -.3], [1, -.35],
nsample=1000)
arma41css = ARMA(y_arma41)
res41css = arma41css.fit(order=(4, 1), trend='nc', method='css')
y_arma14 = arma_generate_sample([1, -.25], [1., -.75, .35, .25, -.3],
nsample=1000)
arma14css = ARMA(y_arma14)
res14css = arma14css.fit(order=(4, 1), trend='nc', method='css')
# ARIMA Model
from statsmodels.datasets import webuse
dta = webuse('wpi1')
wpi = dta['wpi']
mod = ARIMA(wpi, (1, 1, 1)).fit()看完上述內容,是不是對用python進行時間序列分析的方法有進一步的了解,如果還想學習更多內容,歡迎關注億速云行業資訊頻道。
免責聲明:本站發布的內容(圖片、視頻和文字)以原創、轉載和分享為主,文章觀點不代表本網站立場,如果涉及侵權請聯系站長郵箱:is@yisu.com進行舉報,并提供相關證據,一經查實,將立刻刪除涉嫌侵權內容。